Team
Protein methylation dynamics in cancer
Dpt: Signaling through Chromatin
Our research activities
Lysine methylation signaling is a dynamic and reversible process orchestrated by lysine methyltransferases (KMTs, which catalyze the addition of methyl groups), lysine demethylases (KDMs, which remove them), and dedicated methyl-binding proteins that translate these marks into relevant functional outcomes. While methylation of histones is well established as a fundamental regulator of chromatin structure and gene expression, it is increasingly clear that numerous non-histone proteins are also methylated. This suggests that lysine methylation may influence a wide range of cellular processes, and that its deregulation could contribute to various diseases. Despite its biological importance, relatively few non-histone lysine methylation substrates have been identified, and the functional consequences of this signaling remain poorly understood. Given its reversibility, specificity, and druggability, lysine methylation holds high therapeutic potential.
Our research aims to uncover previously uncharacterized KMTs and methylation signaling involved in tumorigenesis and to define novel targetable mechanisms in human cancers. Ultimately, our goal is to establish lysine methylation as a key regulator of cellular homeostasis and to provide new opportunities for therapeutic intervention in cancer.
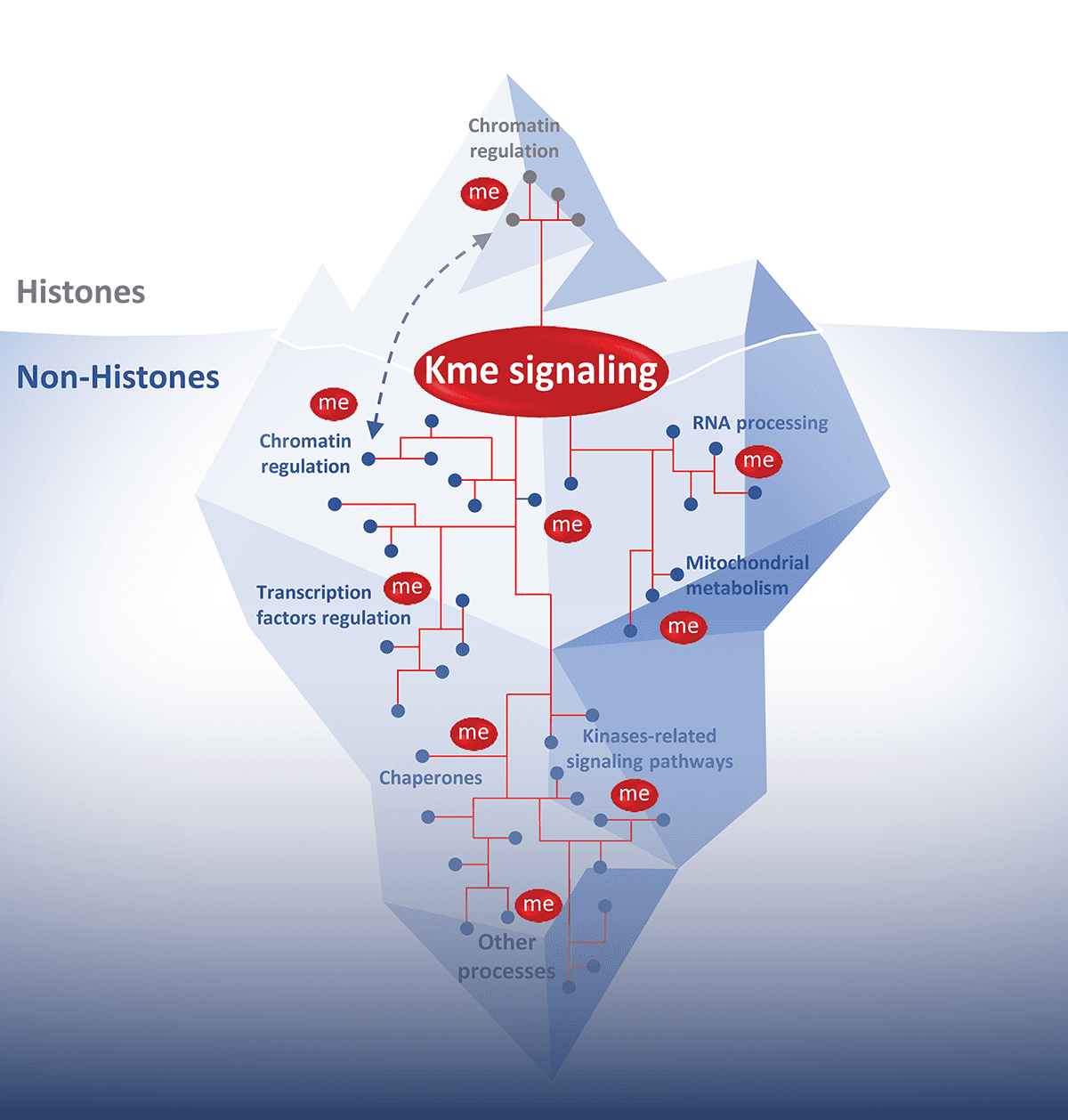
Our research axes
The vast majority of lysine methyltransferases are still poorly characterized and for the most part their activity is unknown or not thoroughly analyzed. Several KMTs are overexpressed or altered in cancer and we aim to characterize key oncogenic lysine methylation signaling.
Learn moreBesides their functions in chromatin regulation, KMTs activities beyond histones modification have rarely been described. We aim to identify key non-histone methylation signaling involved in different biological processes regulating cell homeostasis.
Learn more
Lysine methylation is a finely regulated modification and a dynamic process thanks to demethylases. More than 30 demethylases have been identified in human, and very few have been characterized regarding potential activity on non-histone substrates. We aim to identify the related KDM counteracting key chromatin-independent cellular processes regulated by KMT.
Learn moreOur major publications
See all publicationsOur activities in pictures
Our collaborations
- Mazur lab, MD Anderson Cancer Center, University of Texas, USA
- EDyP Lab, IRIG, France
- Many others !
Our technologies
- High-throughput proteomics
- Cell engineering
- In vitro radiolabeled methylation assay
- Biochemistry and molecular biology tools
- Many more!




