Team
Epigenetics & Chromatin
Dpt: Signaling through Chromatin
Our research activities
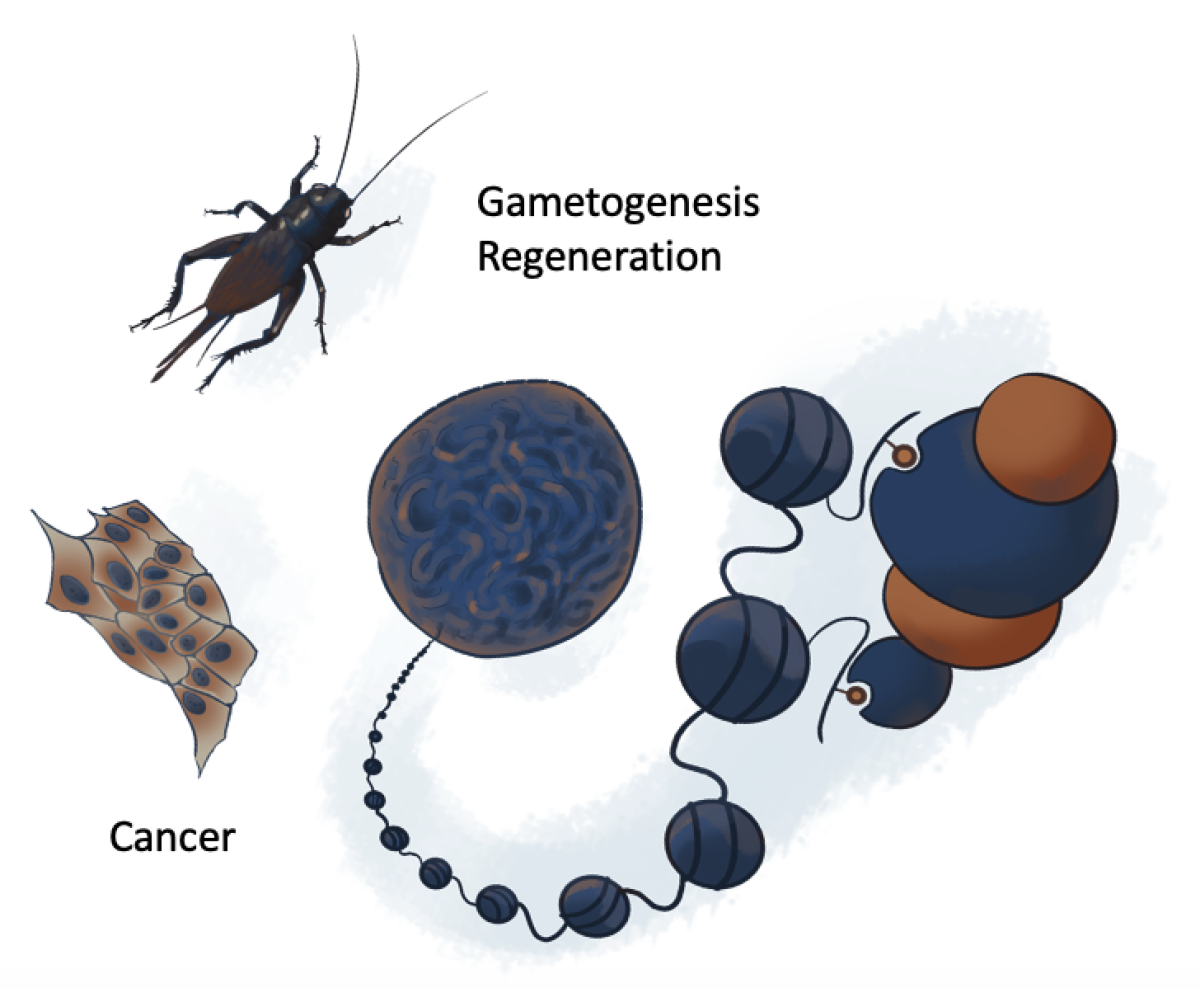
During development, stem cells proliferate and differentiate to compose somatic tissues. By tightly controlling gene expression, chromatin organization is both a defining component of cell identities and a barrier to transcriptional reprogramming that preserves cells from de-differentiation. Yet, following tissue loss or wound, there is a species-specific potential for large-scale regeneration often involving somatic cell regression into a pluripotent state followed by their proliferation and re-differentiation. While powerful to maintain tissue homeostasis, these mechanisms are also dangerous in that they offer a window of opportunity for certain cells to escape their programmed identity. Indeed, cancerous cells often acquire the capacity to overcome and exploit these chromatin barriers to cell transitions to bypass their somatic fate, become proliferative and acquire aberrant identities. Regeneration is therefore a double-edged sword, ensuring tissue integrity but also operating as a backdoor to cancer transformation. Our goal is to understand the epigenetic bases of cell plasticity in regeneration and cancer ultimately seeking to identify actionable mechanisms to facilitate regeneration while avoiding tumorigenesis. Our laboratory studies these mechanisms by implementing high-resolution methods in epigenomics and microscopy focusing on two model cases: hormone dependency in breast cancers and leg regeneration in an insect, the two-spotted cricket Gryllus bimaculatus.
Our research axes
Cancer cells change their epigenome to escape somatic control. Focusing on breast cancer, we want to understand how this adaptation conditions their identity and determines their aggressivity as well as their response to treatments.
As many other insects and arthropods, crickets have the capacity to fully regenerate their legs if amputated. We want to understand the epigenomic mechanisms allowing cells to change their identities several times to ensure this spectacular process goes smoothly.
During spermatozoid differentiation, chromatin undergoes dramatic changes leading to nuclear ultra-compaction. These processes rely on highly orchestrated dynamics of histone modification, replacement, and eviction, and their substitution by sperm-specific proteins notably including protamines. Despite their critical importance, these mechanisms remain a major black box in developmental biology, which we try to elucidate by combining genetics, biochemistry, high-resolution microscopy and biophysical modeling.
Our major publications
See all publicationsOur activities in pictures
Our technologies
- Cut&Run
- MNase-seq
- Stochastic Optical Reconstruction Microscopy (STORM)
- Bioinformatics
- Insect models