Platform
Epinomics
Epigenomics and Transcriptomics
Presentation
Epigenome analysis enables the decoding of dynamic regulations of gene expression.
The Epinomics platform provides expertise in epigenomics and transcriptomics to the academic and industrial scientific communities, applied to basic, translational, and clinical research.
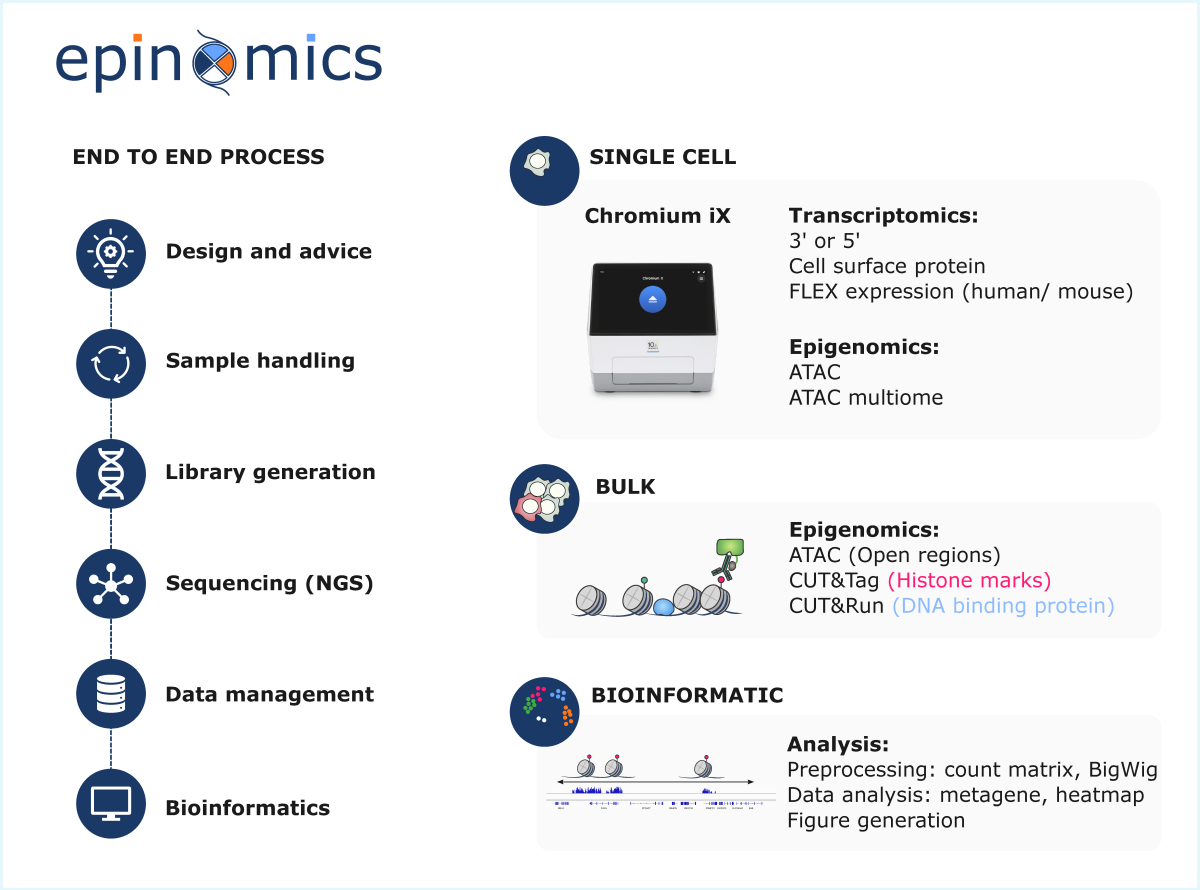
Epinomics performs -omics technologies for studying the epigenome in cell populations (ATAC-Seq, CUT&Tag, CUT&RUN).
The platform provides single-cell analyses of the transcriptome (3’ and 5’ single-cell RNA-Seq) and of chromatin accessibility (single-cell ATAC-Seq), using 10x Genomics technology (starting in 2026). These innovative approaches allow the exploration of cellular heterogeneity, the investigation of rare cell populations, and the study of cell differentiation.
Epinomics further provides expertise in the bioinformatic and statistical analysis of omics data (RNA-Seq, ATAC-Seq, CUT&Tag, CUT&RUN, ChIP-Seq, DNA methylation, Hi-C, etc.). Epinomics also ensures research data management, from data generation to archiving in specialized repositories, following open science best practices, with the objectives of enhancing data visibility and facilitating their citation in scientific publications.
By closely integrating experimental (wet) and bioinformatics (dry) approaches, it offers an end-to-end service covering the entire workflow for chromatin profiling and cellular transcriptomic activity analysis — from sample preparation to data processing and interpretation. The platform also remains open to the development of new technologies, both in bulk and at the single-cell level, to meet emerging needs.
Operational managers
Eve MOUTAUX
Technical manager Epigenomics and Single Cell
Florent CHUFFART
Inserm / IR2, Bioinformatics Technical Manager
Scientific managers
Centers of expertise
Chromatin conformation and histone-associated chemical modifications regulate DNA accessibility in the nucleus and gene expression. Epinomics offers techniques to decipher these regulatory mechanisms in both healthy and pathological tissues:
• ATAC-Seq: Chromatin accessibility
• CUT&Tag: Post-translational modifications of histones
• CUT&RUN: Post-translational modifications of histones or DNA-associated proteins
Epinomics implements 10x Genomics technology for high-throughput analysis of the transcriptome and epigenome at single-cell resolution:
• Single-cell RNA-Seq: 3’ gene expression, 5’ gene expression, FLEX
• Single-cell Epigenomics: ATAC-Seq, Multiomic
Epinomics is also developing plate-based omics technologies for sensitive analysis of rare cell populations. Novel single-cell approaches can be developed in collaboration and tailored to specific scientific needs. Feel free to contact us to discuss!
Epinomics develops expertise and tools dedicated to the management, bioinformatic processing, and statistical analysis of omics data, with a particular focus on transcriptomic and epigenomic domains (RNA-Seq, ATAC-Seq, CUT&Tag, CUT&RUN, ChIP-Seq, DNA methylation, Hi-C).
The platform relies on the high-performance computing infrastructure of the CIMENT/GRICAD mesocenter, on specialized data repositories, and aligns its practices with the principles of open science.
Epinomics is committed to sharing its expertise in epigenomics with the scientific community. The platform offers guidance on choosing appropriate techniques to address specific research questions.
Epinomics supports research teams through pedagogical resources, tutorials, and guidance on the use of bioinformatics tools and computing infrastructures. Epinomics also contributes to teaching in artificial intelligence, machine learning and statistics at university (UGA), for CNRS and for colleagues.



