Équipe
Dynamique de la méthylation des protéines dans le cancer
Dpt: Signalisation et Chromatine
Nos activités de recherche
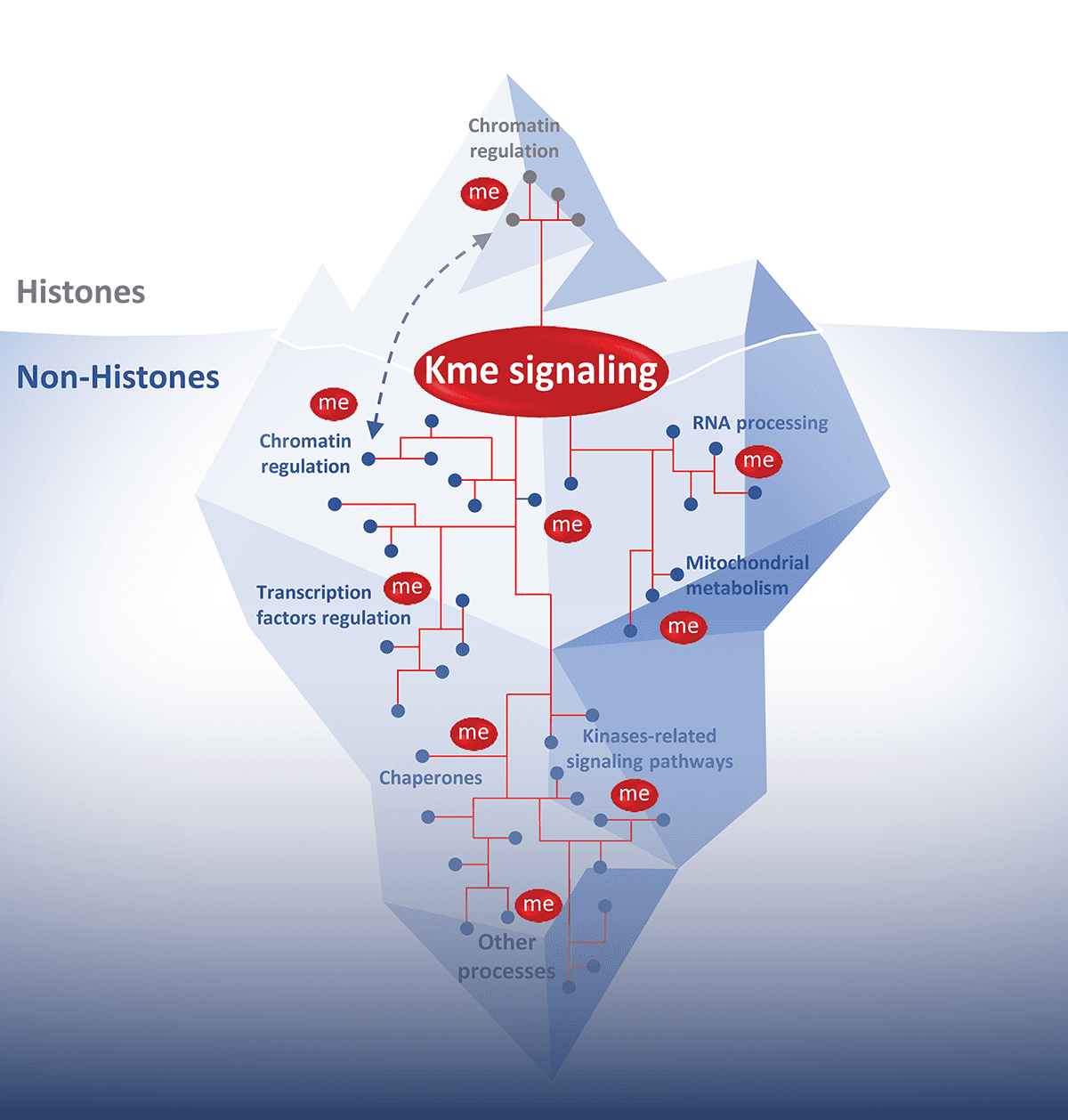
La signalisation par méthylation des lysines est un processus dynamique et réversible, orchestré par les lysine-méthyltransférases (KMTs, qui catalysent l’ajout de groupes méthyle), les lysine déméthylases (KDMs, qui les retirent), ainsi que par des protéines capables de reconnaitre ces marques et les traduire en réponses biologiques pertinentes. Si la méthylation des histones est bien établie comme un régulateur fondamental de la structure de la chromatine et de l’expression des gènes, de nombreuses protéines non-histones sont également méthylées. Cela suggère que la méthylation des lysines pourrait influencer un large éventail de processus cellulaires et que sa dérégulation pourrait contribuer à diverses pathologies. Malgré son importance biologique, relativement peu de substrats non-histones ont été identifiés et les conséquences fonctionnelles de cette signalisation restent encore mal comprises. Compte tenu de sa réversibilité, de sa spécificité et de sa capacité à être ciblée par des inhibiteurs, la méthylation des lysines représente un axe de recherche à fort potentiel thérapeutique.
Nos travaux visent à identifier de nouvelles KMTs et voies de signalisation par méthylation impliquées dans la tumorigenèse, ainsi qu’à définir de nouveaux mécanismes ciblables dans les cancers humains. À terme, notre objectif est d’établir la méthylation des lysines comme un régulateur clé de l’homéostasie cellulaire et d’ouvrir de nouvelles perspectives d’intervention thérapeutique en cancérologie.
Nos axes de recherche
La grande majorité des lysine méthyltransférases sont encore mal caractérisées et pour la plupart leur activité est inconnue. Plusieurs KMTs sont surexprimées ou altérées dans les cancers et nous cherchons à caractériser leurs signalisations oncogéniques.
En savoir plusOutre leurs fonctions dans la régulation de la chromatine, les activités des KMTs au-delà de la modification des histones ont rarement été décrites. Notre objectif est d'identifier les signaux clés de méthylation non-histone impliqués dans différents processus biologiques régulant l'homéostasie cellulaire.
En savoir plus
La méthylation des lysines est une modification finement régulée et un processus rendu dynamique grâce aux déméthylases. Plus de 30 déméthylases ont été identifiées chez l'homme, et très peu ont été caractérisées en ce qui concerne une activité potentielle sur des substrats non histones. Notre objectif est d'identifier les KDMs qui contrent l’action des KMTs non-histones dans les principaux processus cellulaires.
En savoir plusNos publications majeures
Voir toutes les publicationsNos activités en images
Nos collaborations
- Mazur lab, MD Anderson Cancer Center, University of Texas, USA
- EDyP Lab, IRIG, France
- Et beaucoup d’autres !
Nos technologies
- Proteomique haut-débit
- Ingénierie cellulaire
- Test de méthylation radiomarqué in vitro
- Outils de biochimie et biologie moléculaire
- Beaucoup d’autres !




